Dopamine receptors aren’t always dope
The presence of specific allelic variants of the polyamorous dopamine receptor D4 (DRD4) is shown to be predictors of intensified neurodegeneration in frontotemporal dementia-spectrum patients.
Author: Shelby Brock
Download: [ PDF ]
Neuroanatomy
Frontotemporal dementia (FTD) and other neurodegenerative diseases have devastating effects on loved ones and their families. With 50 million global sufferers of dementia,1 the understanding of this and similar neurodegenerative diseases is vital to innovating patient care. However, not much is known about how genotypic to phenotypic variables link in these patients. In a paper published in NeuroImage: Clinical, Butler et al. found two allelic variants of the dopamine receptor D4 (DRD4) associated with increased atrophy in FTD patients.2 This clinically tested link between genotype and atrophy patterns is significant to further the method of FTD diagnosis and potentially develop drugs to target this receptor’s pathways.
Researchers under the grant of the National Institute of Aging (NIA) found reason to hypothesize the known functional polymorphic DRD4 in relation to brain decay based on previously-published studies by Gennatas et al., which cites the polymorphic Val158Met allele variants within the COMT gene, the inhibition of which can alter working memory and decision making.3,4 This study also resembles a 2018 study on the polymorphic effect of DISC1 on dopamine systems as indicators of risk of psychosis in schizophrenia patients.3 The researchers in this study hypothesized that patients with non-wild type variations of the DRD4 receptors would exhibit atrophy in regions where the receptor was most prevalent, which was in frontotemporal regions. DRD4 is a Gcoupled protein which, by inhibition of adenylyl cyclase5 affects the second-messenger system of dopamine, a known deficit which is present in FTD patients.
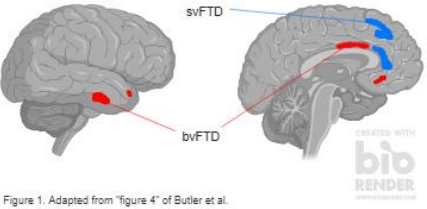
Three groups composed of 337 total participants (healthy controls, Alzheimer’s (AD) patients, and various FTD patients) underwent genomic testing, structural MRI scans, and cognitive testing. The groups were compared using linear modeling of the gray matter maps of healthy controls vs. AD, controls vs. FTD, and AD vs. FTD groups. Genomic testing identified a wild type allelic variant and two variants that dampened the effect of D4, which were hypothesized to exacerbate frontal atrophy. The MRI scans used voxel-based morphometry (VBM) to analyze the intensity of gray matter in brains. Cognitive tests included the CDR sum-of-boxes test, NPI tests, and MMSEs to measure the impairment of function in individuals. Analysis between behavior variant FTD (bvFTD) and semantic variant primary progressive aphasia (svPPA) FTD patients was done using voxelbased morphometry (VBM), which statistically measures and illustrates gray matter structures. All participants were age-matched, had relatively even sex ratios, and education levels, which prevents the interaction of external factors.
After analysis of collected data from participants, the results were not significant for AD patients, but were within FTD patients, with controls and regular symptom-specific atrophy patterns used as comparison. Fifty patients with bvFTD and twenty-six with svPPA and non-wild type variants showed increased gray matter atrophy within the right anterior-inferior insula, dorsal and ventral anterior cingulate cortex, ventromedial prefrontal cortex, and orbitofrontal cortex. The results between the FTD-spectrum patients are illustrated in Figure 1 with a general mapping of the (DRD4) regions where atrophy patterns were amplified beyond syndrome-specific patterns. The hypothesis of DRD4 variant involvement in neurodegeneration was therefore proven correct, but further investigation with more FTD patients was requested by researchers to expand on this correlate.
This study demonstrated the ability to reconcile the link between genetics and behavior as well as addition to scientific literature on the involvement of DRD4 polymorphism on neurodegenerative disease. The identification of DRD4 variants that affect is one step forward in the advancement of patient treatment by emphasizing the need for genomic testing to better perform individualspecific care. The confirmation of receptor involvement in FTD-spectrum atrophy also secondarily aids in the confirmation of FTD (versus a common misdiagnosis of AD). This goes beyond treating an illness; it’s about treating a person. With these findings, more specific drugs can potentially be created to target DRD4 variants known to aggravate atrophy as well as improve current preventative therapy methods.
[+] References
“Dementia.” (12 Dec 2017) World Health Organization, World Health Organization, www.who.int/en/newsroom/fact-sheets/detail/dementia
Butler, P., Chiong, W., Perry, D., Miller, Z., Gennatas, E., Brown, J., Seeley, W. (2019). Dopamine receptor D4 (DRD) polymorphisms with reduced functional potency intensify atrophy in syndrome-specific sites of frontotemporal dementia. NeuroImage: Clinical, 23, 101822. doi: 10.1016/j.nicl.2019.101822
. Farrell, S. M., Tunbridge, E. M., Braeutigam, S., & Harrison, P. J. (2012). COMT Val(158)Met genotype determines the direction of cognitive effects produced by catechol-O-methyltransferase inhibition. Biological psychiatry, 71(6), 538–544. doi:10.1016/j.biopsych.2011.12.023
. Gennatas, E. D., Cholfin, J. A., Zhou, J., Crawford, R. K., Sasaki, D. A., Karydas, A., . . . Seeley, W. W. (2012). COMT Val158Met genotype influences neurodegeneration within dopamine-innervated brain structures. Neurology, 78(21), 1663-1669. doi: 10.1212/wnl.0b013e3182574fa1
DRD4 dopamine receptor D4 [Homo sapiens (human)] - Gene - NCBI. (2019, April 21). Retrieved from https://www.ncbi.nlm.nih.gov/gene/1815
[+] Other Work By Shelby Brock
Effects of neonatal methamphetamine exposure
Neuroscience In Review
The “hunt” for an answer continues
Neurophysiology
Functional deficits have been identified in striatal neurons of mice expressing the mutation for Huntington’s disease. This research identifies a potential target for therapy treatment.