Mini-Brains and Autism: Autism Risk Gene Mutation Leads to Changes in the Cerebellum
A new study finds that removing a single gene associated with autism leads to reduced “mini-brain” size and behavioral impairments.
Author: Emily Dale
Neurophysiology
Introduction
Autism spectrum disorder (ASD) is a complex developmental disorder that presents as a diverse array of behavioral impairments, including altered social cognition and atypical behaviors (McPartland and Volkmar, 2012). ASD is notoriously difficult to study in a laboratory setting, due to the prenatal onset of the disorder, the wide variation in symptoms, and the ever-growing list of genetic and environmental factors thought to be involved in the development of the disorder. These genetic factors, currently thought to be responsible for ASD development present in some patients, and not in others, showing the heterogeneity of this disorder’s developmental cues (McPartland and Volkmar, 2012)(Chaste and Leboyer, 2012). While studies in various models of ASD continue to identify more genes that may be involved in the disorder, it is still unclear what each of these genes is responsible for in both neurotypical and autistic brains.
ASD research is incredibly valuable to the greater population, as the disorder seems to be growing in prevalence each year. It is still unclear whether this disorder is truly becoming more common or if the rise in statistical prevalence is due to improvements to diagnostic criteria and public awareness, but, regardless, people are more likely to be diagnosed with ASD now than ever before (McPartland and Volkmar, 2012)(Chaste and Leboyer, 2012). This heightening in diagnosis calls for further investigation in the causes of the disorder – are there simple behavioral measures that can be taken to avoid ASD development in children, or are the causes strictly genetic?
In a new study, researchers find that one gene linked to autism – the Autism Susceptibility Candidate 2 (AUTS2) gene – has important roles in the initial development of the brain. Specifically, the AUTS2 gene products are necessary for proper cerebellar, or “mini-brain”, development. This study found that knocking out this gene led to improper cerebellar formation and function, as well as behavioral consequences.
The study described here is a seemingly small but vitally important step forward in ASD research. Understanding the biological role of a gene that is commonly associated with ASD and other developmental disorders allows us to draw connections between malfunctioning genes and physical changes in the brain that may lead to disordered thinking.
Background
The Autism susceptibility candidate 2 (AUTS2) gene was originally described as a risk gene for ASD in 2002 (Sultana et al., 2002). Since then, numerous studies have found that this gene has major roles in development of the brain (Hori and Hoshino, 2017)(Monderer-Rothkoff et al., 2021), and mutation of the gene can lead to various developmental and behavioral disorders, including ASD as well as intellectual disability (ID), schizophrenia, ADHD, depression, epilepsy, and others (Hori et al., 2021).
This study focuses on the expression of AUTS2 in the cerebellum, which is a region of the brain primarily credited for the precise coordination of motor movements (Figure 1). However, new studies are showing that the cerebellum also has roles in social cognition and emotional processing, as well as other higher-order cognitive functions (Schmahmann and Caplan, 2006). Interestingly, post-mortem examinations of ASD brains have shown abnormalities in the cerebellum, such as reduced Purkinje cell (a prominent cell type in the cerebellum) size and number (Bauman and Kemper, 2005).
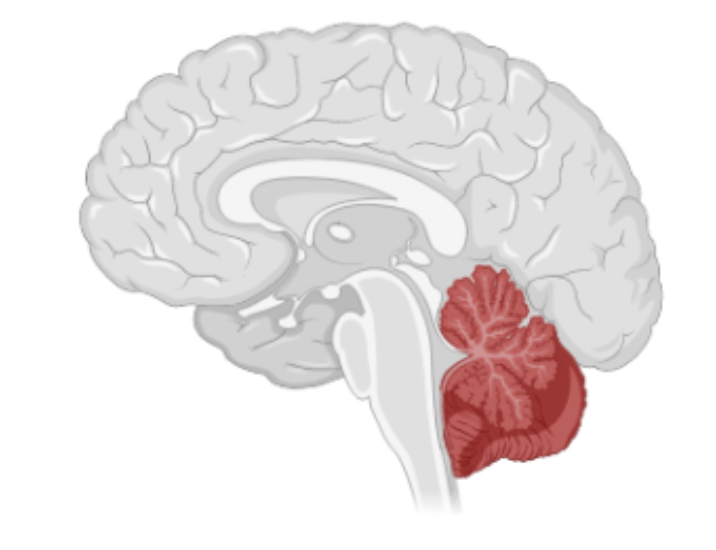
Methods
This study utilized a mouse model of the AUTS2 gene conditional knockout. The mice were bred to express a nonfunctioning AUTS2 gene and were crossed to be homozygous for the knockout. Various tests were performed to quantify differences in brain function and cerebellar structure in the knockout mice compared to controls, which were raised on a similar background and in the same environment to mitigate differences caused by the breeding or raising of the animals.
Immunohistochemistry (IHC) and Nissl staining were used to locate the AUTS2 gene’s expression in cerebellar cells. This technique allowed for easy visual localization of the gene’s expression in brain slices obtained from the knockout mice. Synaptic elements and other cerebellar proteins were also labelled using IHC. Immunoblotting and quantitative PCR (qPCR) was performed to verify levels of expressed AUTS2 in the cerebella in both control and knockout mice. Golgi staining was also conducted in whole brains to quantify dendritic structure and spines.
Electrophysiological measures were conducted on cerebellar slices obtained using a vibratome. Fluorescently labelled Purkinje cells (PCs) in the cerebella were localized and patched for whole-cell recordings. Excitatory post-synaptic currents (EPSCs), miniature EPSCs (mEPSCs), miniature inhibitory post-synaptic currents (mIPSCs), climbing fiber-induced EPSCs (CF-EPSCs), and parallel fiber-induced EPSCs (PF-EPSCs) were all measured using this whole-cell patch recording technique. As necessary to obtain these recordings, several toxins acting at various targets were introduced, including picrotoxin, tetrodotoxin, and NBQX.
Behavioral tests were also conducted on live animals to quantify changes in behavior associated with the AUTS2 knockout. These tests included the rotarod test, a measure of motor coordination (Deacon, 2013), the elevated platform test, a measure of balance and stress response (Degroot et al., 2004), and ultrasonic vocalization recordings which measured communication and courtship behavior of a male mouse when introduced to an unfamiliar female mouse.
Results
Localization investigations in this study revealed that the AUTS2 gene is isolated to two particular cell types in the cerebellum of the mouse – Purkinje cells (PCs), one of the major cell types in the cerebellum, and Golgi cells, which are thought to have a supportive role in cerebellar communication.
The cell types that express this gene were also notably different in morphology than in control animals (Figure 2). In the knockout animals, PCs showed altered, immature-looking dendrite morphology. The connections between PCs and neighboring climbing fibers (CFs) and parallel fibers (PFs) were also unique – the researchers found that there were significantly more synapses altogether. Additionally, there were far fewer PCs overall, making the whole cerebellum smaller in size as compared to controls.
As to be expected with changes in morphology, there were distinct electrophysiological differences seen in the knockout mice brains. The excess of CFs synapses was due to the presence of more CFs innervating each PC (1-3 CFs per PC as compared to 1 CF per PC in controls). The presence of more PF synapses resulted in changes in synaptic activity, as measured by increased EPSC amplitudes and paired pulse ratios.
In behavioral tests, the knockout mice showed signs of impaired motor control and balance, motor learning, and social communication as measured by decreased number and duration of ultrasonic vocalizations.
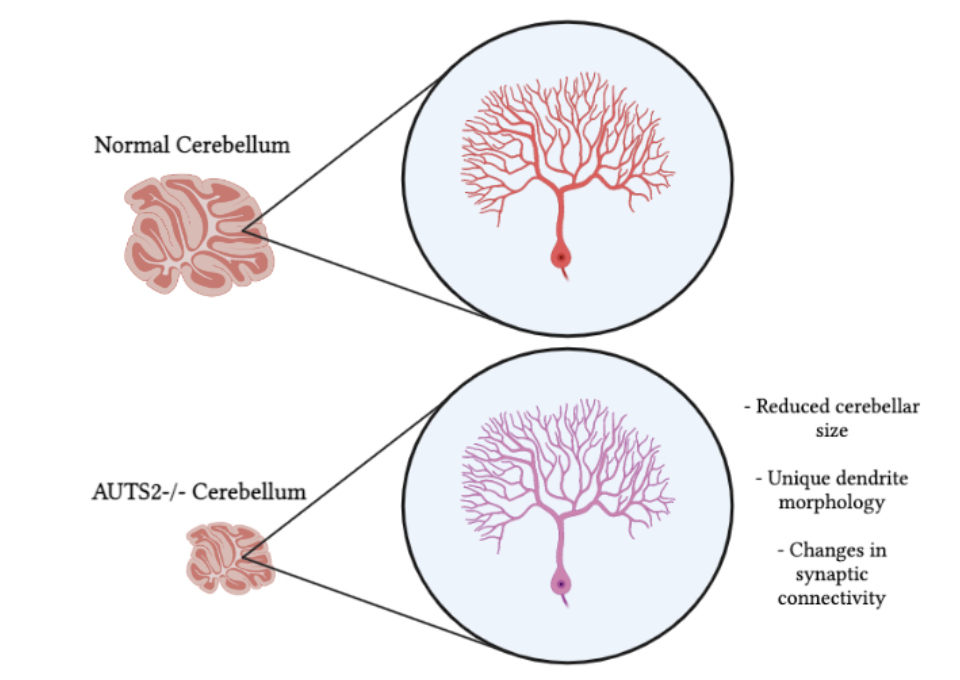
Discussion
The results of this study have large implications in the study of ASD. Although this mutation is not seen in all ASD patients, it has been linked with human ASD patients in past case studies (Sultana et al., 2002) and thus may contribute to the disorder in an unknown number of individuals affected by ASD. In these patients, the altered development of the cerebellum may contribute to the symptoms they face, and changes in connectivity and processing may explain differences in cognition. The deeper understanding of this gene’s role in development may also eventually lead to preventative pharmacology when paired with early pre-natal screening.
Overall, this article is a thorough and enjoyable overview of the AUTS2 gene’s role in cerebellar development and function. This work is incredibly important for the future of ASD research, and hopefully, in the future, other ASD risk genes will be examined with the same scrutiny.
[+] References
Bauman, M. L., & Kemper, T. L. (2005). Neuroanatomic observations of the brain in autism: A review and future directions. International Journal of Developmental Neuroscience: The Official Journal of the International Society for Developmental Neuroscience, 23(2–3), 183–187. https://doi.org/10.1016/j.ijdevneu.2004.09.006
Chaste, P., & Leboyer, M. (2012). Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues in Clinical Neuroscience, 14(3), 281–292.
Deacon, R. M. J. (2013). Measuring Motor Coordination in Mice. Journal of Visualized Experiments : JoVE, 75, 2609. https://doi.org/10.3791/2609
Degroot, A., Wade, M., Salhoff, C., Davis, R. J., Tzavara, E. T., & Nomikos, G. G. (2004). Exposure to an elevated platform increases plasma corticosterone and hippocampal acetylcholine in the rat: Reversal by chlordiazepoxide. European Journal of Pharmacology, 493(1), 103–109. https://doi.org/10.1016/j.ejphar.2004.04.011
Hori, K., & Hoshino, M. (2017). Neuronal Migration and AUTS2 Syndrome. Brain Sciences, 7(5), 54. https://doi.org/10.3390/brainsci7050054
Hori, K., Shimaoka, K., & Hoshino, M. (2021). AUTS2 Gene: Keys to Understanding the Pathogenesis of Neurodevelopmental Disorders. Cells, 11(1), 11. https://doi.org/10.3390/cells11010011
McPartland, J., & Volkmar, F. R. (2012). Autism and Related Disorders. Handbook of Clinical Neurology, 106, 10.1016/B978-0-444-52002-9.00023-1. https://doi.org/10.1016/B978-0-444-52002-9.00023-1
Monderer-Rothkoff, G., Tal, N., Risman, M., Shani, O., Nissim-Rafinia, M., Malki-Feldman, L., Medvedeva, V., Groszer, M., Meshorer, E., & Shifman, S. (2021). AUTS2 isoforms control neuronal differentiation. Molecular Psychiatry, 26(2), 666–681. https://doi.org/10.1038/s41380-019-0409-1
Schmahmann, J. D., & Caplan, D. (2006). Cognition, emotion and the cerebellum. Brain, 129(2), 290–292. https://doi.org/10.1093/brain/awh729
Sultana, R., Yu, C.-E., Yu, J., Munson, J., Chen, D., Hua, W., Estes, A., Cortes, F., de la Barra, F., Yu, D., Haider, S. T., Trask, B. J., Green, E. D., Raskind, W. H., Disteche, C. M., Wijsman, E., Dawson, G., Storm, D. R., Schellenberg, G. D., & Villacres, E. C. (2002). Identification of a Novel Gene on Chromosome 7q11.2 Interrupted by a Translocation Breakpoint in a Pair of Autistic Twins. Genomics, 80(2), 129–134.https://doi.org/10.1006/geno.2002.6810
Yamashiro, K., Hori, K., Lai, E. S. K., Aoki, R., Shimaoka, K., Arimura, N., Egusa, S. F., Sakamoto, A., Abe, M., Sakimura, K., Watanabe, T., Uesaka, N., Kano, M., & Hoshino, M. (2020). AUTS2 Governs Cerebellar Development, Purkinje Cell Maturation, Motor Function and Social Communication. IScience, 23(12). https://doi.org/10.1016/j.isci.2020.101820
[+] Other Work By Emily Dale
Repetitive Transcranial Magnetic Stimulation: Palliative Treatment of the Future?
Neuroanatomy
A new study finds reductions in symptom severity in borderline personality disorder patients after 3 weeks of rTMS on the dorsomedial prefrontal cortical area.
The Roles of Oxidative Stress and Inflammation in Presbycusis
Neuroscience In Review